Chapter 2 Prerequisites
For reproducing the tutorials in the modules 4.2 and 5, we need a few R packages and a Python package. Detailed instructions are below.
2.1 R packages
For module 4.2, we need
reticulateto import the Python package (see next section. You can install it within R:# install reticulate for module 4.2 install.packages("reticulate")For module 5.1, we need
cardelino, which is only available at GitHub, so we need to usedevtools:# install devtools for module 5.1 install.packages('devtools')# install cardelino for module 5.1 devtools::install_github("single-cell-genetics/cardelino", build_vignettes = FALSE)For module 5.2, we need
infercnv, which requires a standalone software JAGS. Please install JAGS manually for Windows or Mac from this link before installrjagsR package: https://sourceforge.net/projects/mcmc-jags/files/JAGS/4.x/
Continue and installrjagsandinfercnv:# install rjags for module 5.2 install.packages('rjags')# install inferCNV for module 5.2 if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager") BiocManager::install("infercnv")
2.2 Python packages
For module 4.2, we need one Python package scvelo. If you haven’t used Python before, we recommend installing it by using Anaconda, which contains Python 3.8 and various useful tools.
2.2.1 Installation on Windows
- Go to Anaconda webpage; click ‘Download’ button to download Anaconda for Windows (with Python3.8).

Double click the downloaded file and install Anaconda following the instructions. (Note, this requires about 3G disk space)
Open
Anaconda Powershell Promptand then create an new conda environment with namesgcellby typingconda create -n sgcell python=3.8Still in the terminal, and activate the environment by typing
conda activate sgcellInstall
scVeloin this conda environment by typing this command in the shell:pip install scvelo==0.2.2
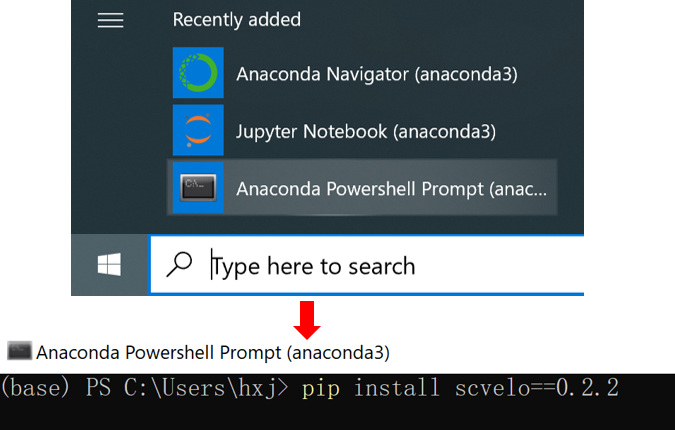
You have successfully set up the base environment of conda and installed the scVelo tool in this environment.
2.2.2 Installation on macOS
In macOS, you could install Anaconda either in graphical mode or command line mode.
Option 1: installation in Graphical Mode (recommended)
This mode is very similar to Installation on Windows.
- Go to Anaconda webpage; click ‘Get Additional Installers’ button and then
select the
64-Bit Graphical Installerfor MacOS.
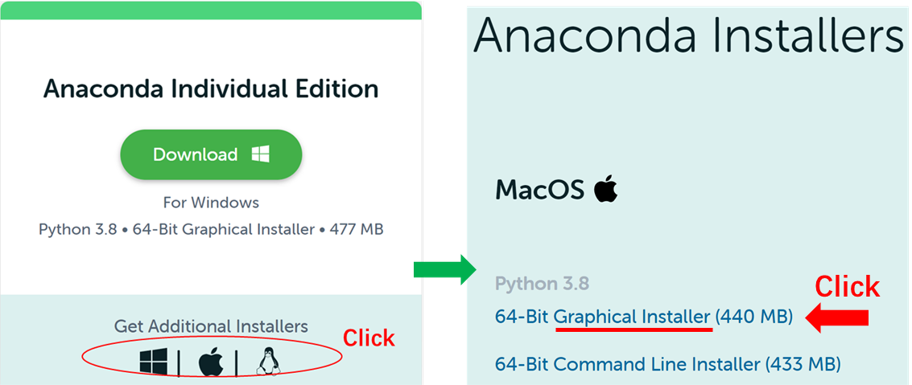
Install Anaconda following the instructions.
Open
Terminal(you can type inSpotlight Search) and then create an new conda environment with namesgcellby typingconda create -n sgcell python=3.8Still in the terminal, and activate the environment by typing
conda activate sgcellInstall
scVeloin this conda environment by typing this command in the shell:pip install scvelo==0.2.2
Option 2: installation in Command Line Mode
This mode is very similar to Installation on Linux.
Open macOS Terminal
Download Anaconda installer with
wget https://repo.anaconda.com/archive/Anaconda3-2021.05-MacOSX-x86_64.shInstall Anaconda with
bash Anaconda3-2021.05-MacOSX-x86_64.sh; follow the prompts on the installer screens.
When you are askedDo you wish the installer to initialize Anaconda3 by running conda init, we recommend “yes.”Close and then re-open your Shell, to make the changes take effect.
Create an new conda environment with name
sgcellby typingconda create -n sgcell python=3.8Still in the terminal, and activate the environment by typing
conda activate sgcellInstall
scVeloin this conda environment by typing this command in the shell:pip install scvelo==0.2.2
2.2.3 Installation on Linux
Open Linux Shell.
Download Anaconda installer with
wget https://repo.anaconda.com/archive/Anaconda3-2021.05-Linux-x86_64.shInstall Anaconda with
bash Anaconda3-2021.05-Linux-x86_64.sh; follow the prompts on the installer screens.
When you are askedDo you wish the installer to initialize Anaconda3 by running conda init, we recommend “yes.”Close and then re-open your Shell, to make the changes take effect.
Create an new conda environment with name
sgcellby typingconda create -n sgcell python=3.8Still in the terminal, and activate the environment by typing
conda activate sgcellInstall
scVeloin this conda environment by typing this command in the shell:pip install scvelo==0.2.2Type
conda listin Shell.
A list of installed packages appears if it has been installed correctly.